Recommended Papers to Read
- J. I. Joo, H.-J. Park, and K.-H. Cho*, “Normalizing input-output relationships of cancer networks for reversion therapy”, Advanced Science, 2207322, pp. 1-13, Jun. 2023. [pdf]
- D. Shin and K.-H. Cho*, “Critical Transition and Reversion of Tumorigenesis”, Experimental & Molecular Medicine, 55, pp. 692-705, Apr. 2023. [pdf]
- N. Kim, C. Y. Hwang, T. Kim, H. Kim, and K.-H. Cho*, “A cell fate reprogramming strategy reverses epithelial-to-mesenchymal transition of lung cancer cells while avoiding hybrid states”, Cancer Research, 83, 6, pp. 956-970, Mar. 2023. [pdf]
- S.R. Choi, C.Y. Hwang, J. Lee, and K.-H. Cho*, “Network analysis identifies regulators of basal-like breast cancer reprogramming and endocrine therapy vulnerability”, Cancer Research, 82, 2, pp. 320-330, Jan. 2022. [pdf]
- J.-C. Park, S.-Y. Jang, D. Lee, J. Lee, U. Kang, H. Chang, H. J. Kim, S.-H. Han, J. Seo, M. Choi, D. Y. Lee, M. S. Byun, D. Yi, K.-H. Cho*, I. Mook-Jung*, “A logical network-based drug-screening platform for Alzheimer’s disease representing pathological features of human brain organoids”, Nature Communications, 12, 280, pp.1-13, Jan. 2021. [pdf]
- S. An, S.-Y. Cho, J. Kang, S. Lee, H.-S. Kim, D.-J. Min, E. Son, and K.-H. Cho*, “Inhibition of 3-phosphoinositide–dependent protein kinase 1 (PDK1) can revert cellular senescence in human dermal fibroblasts”, PNAS (Proceedings of the National Academy of Sciences of the United States of America), 117, 49, pp. 31535-31546, Dec. 2020. [pdf]
- S.-M. Park, C. Y. Hwang, J. Choi, C. Y. Joung, and K.-H. Cho*, “Feedback analysis identifies a combination target for overcoming adaptive resistance to targeted cancer therapy”, Oncogene, 39, 19, pp. 3803-3820, May 2020. [pdf]
- S. Lee, C. Lee, C. Y. Hwang, D. Kim, Y. Han, S. N. Hong, S.-H. Kim, and K.-H. Cho*, “Network inference analysis identifies SETDB1 as a key regulator for reverting colorectal cancer cells into differentiated normal-like cells”, Molecular Cancer Research, 18, 1, pp.118-129, Jan. 2020. [pdf]
- D. Lee and K.-H. Cho*, “Signal flow control of complex signaling networks”, Scientific Reports, 9, 14289, pp.1-18, Oct. 2019. [pdf]
- S.-M. Choo, S.-M. Park, and K.-H. Cho*, “Minimal intervening control of biomolecular networks leading to a desired cellular state”, Scientific Reports, 9, 13124, pp.1-15, Sep. 2019. [pdf]
- B. Lee, U. Kang, H. Chang, and K.-H. Cho*, “The hidden control architecture of complex brain networks”, iScience (Cell Press), 13, pp.154-162, Mar. 2019. [pdf]
- B. Lee, D. Shin, S. P. Gross, and K.-H. Cho*, “Combined positive and negative feedback allows modulation of neuronal oscillation frequency during sensory processing”, Cell Reports, 25, 6, pp.1548-1560, Nov. 2018. [pdf]
- S.-Y. Yeo, K.-W. Lee, D. Shin, S. An, K.-H. Cho*, and S.-H. Kim, “A positive feedback loop bi-stably activates fibroblasts”, Nature Communications, 9, 3016, pp.1-16, Aug. 2018. [pdf]
- S.-M. Choo, B. Ban, J. I. Joo, and K.-H. Cho*, “The phenotype control kernel of a biomolecular regulatory network”, BMC Systems Biology, 12, 49, pp.1-15, Apr. 2018. [pdf]
- M. Choi, J. Shi, Y. Zhu, R. Yang, and K.-H. Cho*, “Network dynamics-based stratification of cancer panel for systemic prediction of anticancer drug response”, Nature Communications, 8, 1940, pp. 1-12, Dec. 2017. [pdf]
- D. Shin, J. Lee, J.-R. Gong, and K.-H. Cho*, “Percolation transition of cooperative mutational effects in colorectal tumorigenesis”, Nature Communications, 8, 1270, pp.1-14, Nov. 2017. [pdf]
- K.-H. Cho*, S. Lee, D. Kim, D. Shin, J. I. Joo, and S.-M. Park, “Cancer reversion, a renewed challenge in systems biology”, Current Opinion in Systems Biology, 2, pp. 48-57, Apr. 2017. [pdf]
- K.-H. Cho*, J. I. Joo, D. Shin, D. Kim, and S.-M. Park, “The reverse control of irreversible biological processes”, WIREs Systems Biology and Medicine, 8, 5, pp. 366-377, Sep/Oct. 2016. [pdf]
- T.-H. Kim, N. Monsefi, J.-H. Song, A. von Kriegsheim, D. Vandamme, O. Pertz, B. N. Kholodenko, W. Kolch*, and K.-H. Cho*, “Network based identification of feedback modules that control RhoA activity and cell migration”, Journal of Molecular Cell Biology, Vol. 7, Issue 3, pp. 242-252, Jun. 2015. [pdf]
- S.-Y. Shin, T. Kim, H.-S. Lee, J. H. Kang, J. Y. Lee, K.-H. Cho* and D. H. Kim, “The switching role of β-adrenergic receptor signalling in cell survival or death decision of cardiomyocytes”, Nature Communications, Vol. 5:5777, pp. 1-13, Dec. 2014. [pdf]
- D. Shin, I. S. Kim, J. M. Lee, S.-Y. Shin, J.-H. Lee, S. H. Baek*, and K.-H. Cho*, “The hidden switches underlying RORα-mediated circuits that critically regulate uncontrolled cell proliferation”, Journal of Molecular Cell Biology, Vol. 6, Issue 4, pp. 338-348, Aug. 2014. [pdf]
- J.-H. Song, D. J. Huels, R. A. Ridgway, O. J. Sansom, B. N. Kholodenko, W. Kolch, and K.-H. Cho*, “The APC network regulates the removal of mutated cells from colonic crypts”, Cell Reports (a new Cell press journal), Vol. 7, Issue 1, pp. 94-103, Apr. 2014. [pdf]
- D. Shin and K.-H. Cho*, “Recurrent connections form a phase-locking neuronal tuner for frequency-dependent selective communication”, Scientific Reports (Nature Publishing Group), Vol. 3: 2519, pp. 1-8, Aug. 2013. [pdf]
- J. Kim, S.-M. Park, and K.-H. Cho*, “Discovery of a kernel for controlling biomolecular regulatory networks”, Scientific Reports (Nature Publishing Group), Vol. 3: 2223, pp. 1-9, Jul. 2013. [pdf]
- M. Choi, J. Shi, S. H. Jung, X. Chen, and K.-H. Cho*, “Attractor Landscape Analysis Reveals Feedback Loops in the p53 Network That Control the Cellular Response to DNA Damage”, Science Signaling, Vol. 5, Issue 251, ra83, Nov. 2012. [pdf]
- J.-K. Won, H. W. Yang, S.-Y. Shin, J. H. Lee, W. D. Heo*, and K.-H. Cho*, “The Cross Regulation Between ERK and PI3K Signaling Pathways Determines the Tumoricidal Efficacy of MEK Inhibitor”, Journal of Molecular Cell Biology, Vol. 4, Issue 3, pp. 153-163, Jun. 2012. [pdf]
- J.-R. Kim, J. Kim, Y.-K. Kwon, H.-Y. Lee, P. Heslop-Harrison, and K.-H. Cho*, “Reduction of Complex Signaling Networks to a Representative Kernel”, Science Signaling, Vol. 4, Issue 175, ra35, May 2011. [pdf]
- S.-Y. Shin, O. Rath, A. Zebisch, S.-M. Choo, W. Kolch, and K.-H. Cho*, “Functional roles of multiple feedback loops in ERK and Wnt signaling pathways that regulate epithelial-mesenchymal transition”, Cancer Research, Vol. 70, Issue 17, pp. 6715-6724, Sep. 2010. [pdf]
International Journals
- Y. Kim, Y. Han, C. Hopper, J. Lee, J. I. Joo, J.-R. Gong, C.-K. Lee, S.-H. Jang, J. Kang, T. Kim, and K.-H. Cho*, “A grey box framework that optimizes a white box logical model using a black box optimizer for simulating cellular responses to perturbations”, Accepted for publication in Cell Reports Methods, Apr. 2024.
- J. Lee, N. Kim, and K.-H. Cho*, “Decoding the principle of cell-fate determination for its reverse control”, Accepted for publication in npj Systems Biology and Applications, Apr. 2024.
- Y. Kim, S. R. Choi, and K.-H. Cho*, “Reducing state conflicts between network motifs synergistically enhances cancer drug effects and overcomes adaptive resistance”, Cancers, 16(7), 1337, Mar. 2024. [Abstract]
- D. Kim, C. Y. Hwang, and K.-H. Cho*, “The fitness trade-off between growth and stress resistance determines the phenotypic landscape”, BMC Biology, 22, 62, Mar. 2024. [Abstract]
- S. M. Lee, Y. Han, and K.-H. Cho*, “Deep learning untangles the resistance mechanism of p53 reactivator in lung cancer cells”, iScience, 26, 108377, Dec. 2023. [Abstract]
- J. I. Joo, H.-J. Park, and K.-H. Cho*, “Normalizing input-output relationships of cancer networks for reversion therapy”, Advanced Science, 2207322, pp. 1-13, Jun. 2023.

[Abstract]
“Inside Back Cover Paper – A genetic alteration can disrupt input-output relationship of the intracellular regulatory network within a cancer cell. However, by making use of the inherent redundancy remained in the complex network, we can still restore the input-output relationship through regulation of a proper molecular target and induce cancer reversion. More details can be found in article number 2207322 by Jae Il Joo, Hwa-Jeong Park, and Kwang-Hyun Cho.” - J. Kim, C. Hopper, and K.-H. Cho*, “Statistical control of structural networks with limited interventions to minimize cellular phenotypic diversity represented by point attractors”, Scientific Reports, 13, 6275, pp.1-15, Apr. 2023. [Abstract]
- K.-H. Lee*, S. R. Yoon, J.-R. Gong, E.-J. Choi, H. S. Kim, C.-J. Park, S.-C. Yun, S.-Y. Park, S.-J. Jung, H. Kim, S. Y. Lee, H. Jung, J.-E. Byun, M. Kim, S.-Y. Kim, J.-H. Kim, J.-H. Lee, J.-H. Lee, Y. Choi, H.-S. Park, Y.-S. Lee, Y.-A. Kang, M. Jeon, J. M. Woo, H. Kang, S. Baek, S. M. Kim, H.-M. Kim, K.-H. Cho*, and I. Choi*, “The Infusion of Ex Vivo, Interleukin-15 and -21-Activated Donor NK Cells after Haploidentical HCT in High-Risk AML and MDS Patients – A Randomized Trial”, Leukemia, 37, pp. 807-819, Mar. 2023. [Abstract]
- S. An, S-Y. Jang, S.-M. Park, C.-K. Lee, H.-M. Kim, and K.-H. Cho*, “Global stabilizing control of large-scale biomolecular regulatory networks”, Bioinformatics, 39, 1, Jan. 2023. [Abstract]
- D. Shin and K.-H. Cho*, “Critical Transition and Reversion of Tumorigenesis”, Experimental & Molecular Medicine, 55, pp. 692-705, Apr. 2023. [Abstract]
- N. Kim, C. Y. Hwang, T. Kim, H. Kim, and K.-H. Cho*, “A cell fate reprogramming strategy reverses epithelial-to-mesenchymal transition of lung cancer cells while avoiding hybrid states”, Cancer Research, 83, 6, pp. 956-970, Mar. 2023.

[Abstract]
“This paper was selected as a cover paper of March issue in 2023 – The epithelial-to-mesenchymal transition (EMT) of primary cancer cells contributes to the acquisition of lethal properties by tumors, including metastasis and drug resistance. A cell-fate reprogramming strategy based on attractor landscapes can unravel various plastic routes of EMT and discover potential therapeutic targets for EMT reversal while avoiding hybrid states.” - M. Choi, S.-M. Park, and K.-H. Cho*, “Evaluating a therapeutic window for precision medicine by integrating genomic profiles and p53 network dynamics”, Communications Biology, 5, 924, pp.1-13, Sep. 2022. [Abstract]
- J. Lee, S. R. Choi, and K.-H. Cho*, “Network dynamics caused by genomic alteration determine the therapeutic response to FGFR inhibitors for lung cancer”, Biomolecules, 12, 1197, pp.1-14, Aug. 2022. [Abstract]
- D. Kim and K.-H. Cho*, “Hidden patterns of gene expression provide prognostic insight for colorectal cancer”, Cancer Gene Therapy, pp.1-11, Aug. 2022. [Abstract]
- J.-M. Yang, C.-K. Lee, and K.-H. Cho*, “Robust stabilizing control of perturbed biological networks via coordinate transformation and algebraic analysis”, Accepted for publication in IEEE Transactions on Neural Networks and Learning Systems, July 2022. [Abstract]
- J.-C. Park, N. Barahona-Torres, S.-Y. Jang, K. Y. Mok, H. J. Kim, S.-H. Han, K.-H. Cho, X. Zhou, A. K. Y. Fu, N. Y. Ip, J. Seo, M. Choi, H. Jeong, D. Hwang, D. Y. Lee, M. S. Byun, D. Yi, J. W. Han, I. Mook-Jung, and J. Hardy, “Multi-omics-based autophagy-related untypical subtypes in patients with cerebral amyloid pathology”, Advanced Science, 2201212, pp.1-17, June 2022. [Abstract]
- K.-W. Lee, S.-Y. Yeo, J.-R. Gong, O.-J. Koo, I. Sohn, W. Y. Lee, H. C. Kim, S. H. Yun, Y. B. Cho, M.-A. Choi, S. An, J. Kim, C. O. Sung*, K.-H. Cho*, S.-H. Kim*, “PRRX1 is a master transcription factor of stromal fibroblasts for myofibroblastic lineage progression”, Nature Communications, 13, 2793, pp.1-23, May 2022. [Abstract]
- B. Lee, U. Kang, H. Chang, and K.-H. Cho*, “The hidden community architecture of human brain networks”, Scientific Reports, 12, 3540, pp.1-8, Mar. 2022. [Abstract]
- N. Shakiba.C, Li.J. Garcia-Ojalvo, K.-H. Cho, K. Patil, A. Walczak, Y.-Y. Liu, S. Kuehn, Q. Nie, A. Klein, G. Deco, M. Kringelbach, S. Iyer-Biswas, “How can Waddington-like landscapes facilitate insights beyond developmental biology?”, Cell Systems, 13, 1, pp. 4-9, Jan. 2022. [Link]
- S.R. Choi, C.Y. Hwang, J. Lee, and K.-H. Cho*, “Network analysis identifies regulators of basal-like breast cancer reprogramming and endocrine therapy vulnerability”, Cancer Research, 82, 2, pp. 320-330, Jan. 2022. [Abstract]
- S.-J. Park and K.-H. Cho*, “Discrete event dynamic modeling and analysis of the democratic progress in a society controlled by network agents”, IEEE Transactions on Automatic Control, 67, 1, pp. 359-365, Jan. 2022. [Abstract]
- J.-M. Yang, C.-K. Lee, and K.-H. Cho*, “Stabilizing control of complex biological networks based on attractor-specific network reduction”, IEEE Transactions on Control of Network Systems, 8, 2, pp. 928-939, Jun. 2021. [Abstract]
- C. Y. Hwang, S. J. Yu, J.-K. Won, S.-M. Park, H. Noh, S. Lee, E. J. Cho, J.-H. Lee, K. B. Lee, Y. J. Kim, K.-S. Suh, J.-H. Yoon, and K.-H. Cho*, “Systems analysis identifies endothelin 1 axis blockade for enhancing the anti-tumor effect of multikinase inhibitor”, Cancer Gene Therapy, 29, pp. 845-858, Aug. 2021. [Abstract]
- H.-Y. Lee, Y. Jeon, Y. K. Kim, J. Y. Jang, Y. S. Cho, J. Bhak*, K.-H. Cho*, “Identifying molecular targets for reverse aging using integrated network analysis of transcriptomic and epigenomic changes during aging”, Scientific Reports, 11, 12317, pp.1-13, Jun. 2021. [Abstract]
- K.-H. Cho*, S. An, J. Kang, “Systems biology for reverse aging”, Aging, Jun. 2021. [Abstract]
- I. Yang and K.-H. Cho*, “A low-power timing-error tolerant circuit by controlling a clock”, IEEE Transactions on Very Large Scale Integration Systems, 29, 3, pp.512-518, Mar. 2021. [Abstract]
- J.-C. Park, S.-Y. Jang, D. Lee, J. Lee, U. Kang, H. Chang, H. J. Kim, S.-H. Han, J. Seo, M. Choi, D. Y. Lee, M. S. Byun, D. Yi, K.-H. Cho*, I. Mook-Jung*, “A logical network-based drug-screening platform for Alzheimer’s disease representing pathological features of human brain organoids”, Nature Communications, 12, 280, pp.1-13, Jan. 2021. [Abstract]
“Selected as Nature Communications Editors’ highlights (https://www.nature.com/collections/ecgchegdh) – Developing effective drugs for Alzheimer’s disease (AD), the most common cause of dementia, has been difficult because of complicated pathogenesis. Here, the authors report an efficient network-based drug-screening platform developed by integrating mathematical modeling and the pathological features of human cerebral organoids.” - S. An, S.-Y. Cho, J. Kang, S. Lee, H.-S. Kim, D.-J. Min, E. Son, and K.-H. Cho*, “Inhibition of 3-phosphoinositide–dependent protein kinase 1 (PDK1) can revert cellular senescence in human dermal fibroblasts”, PNAS (Proceedings of the National Academy of Sciences of the United States of America), 117, 49, pp. 31535-31546, Dec. 2020. [Abstract]
- S.-M. Choo, L. M. Almomani, and K.-H. Cho*, “Boolean feedforward neural network modeling of molecular regulatory networks for cellular state conversion”, Frontiers in Physiology, 11, 594151, pp. 1-14, Dec. 2020. [Abstract]
- J. Choi, J.-R. Gong, C. Y. Hwang, C. Y. Joung, S. Lee and K.-H. Cho*, “A systems biology approach to identifying a master regulator that can transform the fast growing cellular state to a slowly growing one in early colorectal cancer development model”, Frontiers in Genetics (Special Issue on Dynamics and Complexity of Molecular Netowrks Controlling Cell Fate Decisions), 11, 570546, pp. 1-13, Oct. 2020. [Abstract]
- H.-S. Lee, S. Lee, and K.-H. Cho*, “Cotargeting BET proteins overcomes resistance arising from PI3K/mTOR blockade-induced pro-tumorigenic senescence in colorectal cancer”, International Journal of Cancer, 147, 10, pp. 2824-2837, Nov. 2020. [Abstract]
“Selected as a Featured Article (https://onlinelibrary.wiley.com/doi/10.1002/ijc.33142) – Targeting initially a different signaling pathway, Lee et al (“Cotargeting BET proteins overcomes resistance arising from PI3K/mTOR blockade‐induced pro‐tumorigenic senescence in colorectal cancer,” doi: 10.1002/ijc.33047) observed that colorectal cancer (CRC) cells become resistant to inhibitors of the PI3K/mTOR pathway, one of the pathways that is frequently deregulated in CRC, through feed‐back activation of multiple receptor tyrosine kinases (RTKs) and, consequently, of their downstream signaling. At the same time, p38 stress kinase was activated and induced cellular senescence, which in turn led to the secretion of cytokines supporting migration and invasion. The authors decided to try and block activated transcription at the chromatin level with the BET inhibitor JQ1. Notably, the addition of JQ1 dramatically suppressed cell growth, and highly increased cell death compared to the treatment of either drug alone. JQ1 reduced the expression levels of EGFR and IGF1R and also counteracted the induction of IL‐8 and MIF that were increased by PI3K/mTOR inhibition. These data suggest that upregulation of the RTKs and cytokines may be BET‐dependent. The authors support the relevance of their observations by xenograft experiments, which confirm the efficiency of the combination treatment, and by data extracted from The Cancer Genome Atlas, which indicate that mRNA levels of BRD4 transcripts are significantly elevated in colorectal tumors when compared with normal matched tissues and that high BRD4 levels correlate with a poor clinical outcome.” - J. I. Joo, M. Choi, S.-H. Jang, S. Choi, S.-M. Park, D. Shin, and K.-H. Cho*, “Realizing cancer precision medicine by integrating systems biology and nanomaterial engineering”, Advanced Materials (IF: 27.398), 32, 35/1906783, pp. 1-23, Apr. 2020.
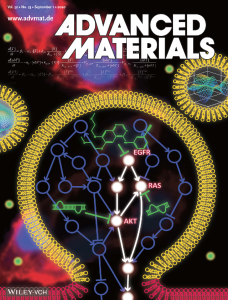
[Abstract]
“Inside Back Cover (https://onlinelibrary.wiley.com/doi/abs/10.1002/adma.202070265) – the cover caption : Systems‐biology can identify novel drug targets through computational analysis of cellular regulatory networks, and nanomaterial engineering can develop nanocarriers to deliver therapeutic agents to the specific target of cancer cells. In article number 1906783, Kwang‐Hyun Cho and co‐workers review the recent progress and challenges in systems biology and nanomedicine and their integration with a view toward cancer precision medicine.” - S.-M. Park, C. Y. Hwang, J. Choi, C. Y. Joung, and K.-H. Cho*, “Feedback analysis identifies a combination target for overcoming adaptive resistance to targeted cancer therapy”, Oncogene, 39, 19, pp. 3803-3820, Mar. 2020. [Abstract]
- S.-J. Park and K.-H. Cho*, “Achieving a global objective with competing networked agents in the framework of discrete event systems”, International Journal of Control, 93, 4, pp. 889-897, Mar. 2020. [Abstract]
- S. Lee, C. Lee, C. Y. Hwang, D. Kim, Y. Han, S. N. Hong, S.-H. Kim, and K.-H. Cho*, “Network inference analysis identifies SETDB1 as a key regulator for reverting colorectal cancer cells into differentiated normal-like cells”, Molecular Cancer Research, 18, 1, pp.118-129, Jan. 2020.

[Abstract]
“Featured Cover Article (https://mcr.aacrjournals.org/content/18/1.cover-expansion) – the cover caption : During the process of tumorigenesis, differentiated cells progressively acquire changes to their gene expression program that drive the cell toward a malignant, stem cell-like state. The precise mechanisms underlying these changes are poorly defined, though it can generally be said that de-differentiation primarily occurs through the loss or inactivation of cell lineage factors. In this issue, Lee and colleagues detail new insights into the molecular mechanisms regulating the differentiation states of colorectal cancer cells. They further suggest possible avenues to reprogram tumor cells into a nonmalignant post-mitotic state by overcoming the epigenetic barrier to the differentiated state, as depicted conceptually on the cover. [Image: Kwang-Hyun Cho, Korea Advanced Institute of Science and Technology]”
“Highlight in Molecular Cancer Research, January 2020
(https://mcr.aacrjournals.org/content/18/1/1) : Reverting Colorectal Cancer Cells to Normal-like Cells – Genetic and epigenetic alterations progressively rewire gene expression networks in tumor cells, leading to the emergence of malignant phenotypes. Here, Lee and colleagues demonstrate the feasibility of reversing colorectal cancer cells to post-mitotic, normal-like cells via an epigenetic mechanism. To identify transcription factors (TF) that control the differentiated state, the authors constructed a gene regulatory network using transcriptomic data from normal colon mucosa. Network analysis identified the histone methyltransferase SETDB1 as a key disruptor of the normal differentiation: expression of SETDB1 impeded the function of key lineage-determining TFs, thus undermining the differentiated state. Accordingly, depletion of SETDB1 from colorectal cancer cells restored lineage TF function and reverted the cellular transcriptome back to a normal-like state. Taken together, the data provide proof of concept for a new therapeutic method that returns cancer cells to a non-malignant state rather than seeking to eliminate them through cytotoxic modalities.“ - D. Woo, Y. Seo, N. Kim, H. Jung, S.-M. Park, S. Lee, H. Lee, K.-H. Cho*, W. D. Heo*, “Locally activating TrkB receptor generates actin waves and specifies axonal fate”, Cell Chemical Biology, 26, 12, pp. 1-12, Dec. 2019.

[Abstract]
“This paper was selected as a cover paper of December issue in 2019.”
“This paper was previewed by Michael Z. Lin in Cell Chemical Biology: ”
“(https://www.sciencedirect.com/science/article/pii/S245194561930399X)” - D. Lee and K.-H. Cho*, “Signal flow control of complex signaling networks”, Scientific Reports, 9, 14289, pp.1-18, Oct. 2019. [Abstract]
- S.-M. Choo, S.-M. Park, and K.-H. Cho*, “Minimal intervening control of biomolecular networks leading to a desired cellular state”, Scientific Reports, 9, 13124, pp.1-15, Sep. 2019. [Abstract]
- S. Shin, M.-W. Kim, K.-H. Cho*, and L. Nguyen, “Coupled feedback regulation of nuclear factor of activated T-cells (NFAT) modulates activation-induced cell death of T cells”, Scientific Reports, 9, 10637, pp.1-15, Jul. 2019. [Abstract]
- M. P. Menden et al. (K.-H. Cho, S. Cho, Y. Han, J. Kim, Y. Kim, J.-H. Song as members of AstraZeneca-Sanger Drug Combination DREAM Consortium, “Community assessment to advance computational prediction of cancer drug combinations in a pharmacogenomic screen”, Nature Communications, 10, 2674, pp. 1-17, Jun. 2019. [Abstract]
- I. Yang and K.-H. Cho*, “Transient-error correction system with real-time logic switching inspired from attractor-conversion characteristics of a cancer cell”, Microelectronics Reliability, 96, pp.51-59, May. 2019. [Abstract]
- S.-M. Park, C. Y. Hwang, S.-H. Cho, D. Lee, J.-R. Gong, S. Lee, S. Nam, and K.-H. Cho*, “Systems analysis identifies potential target genes to overcome cetuximab resistance in colorectal cancer cells”, The FEBS Journal, 286, 7, pp.1305-1318, Apr. 2019.

[Abstract]
“This paper was selected as a cover paper of April issue in 2019.”
“A Commentary on this paper was published in The FEBS Journal (https://doi.org/10.1111/febs.14809).” - B. Lee, U. Kang, H. Chang, and K.-H. Cho*, “The hidden control architecture of complex brain networks”, iScience (Cell Press), 13, pp.154-162, Mar. 2019. [Abstract]
- B. Lee, D. Shin, S. P. Gross, and K.-H. Cho*, “Combined positive and negative feedback allows modulation of neuronal oscillation frequency during sensory processing”, Cell Reports, 25, 6, pp.1548-1560, Nov. 2018. [Abstract]
- J.T. Kim, S.H. Jung, and K.-H. Cho*, “Efficient harmonic peak detection of vowel sounds for enhanced voice activity detection”, IET Signal Processing, 12, 8, pp.975-982, Oct. 2018. [Abstract]
- J. I. Joo, J. X. Zhou, S. Huang, and K.-H. Cho*, “Determining relative dynamic stability of cell states using Boolean network model”, Scientific Reports, 8, 12077, pp.1-14, Aug. 2018. [Abstract]
- S.-Y. Yeo, K.-W. Lee, D. Shin, S. An, K.-H. Cho*, and S.-H. Kim, “A positive feedback loop bi-stably activates fibroblasts”, Nature Communications, 9, 3016, pp.1-16, Aug. 2018. [Abstract]
- J.-M. Yang, C.-K. Lee, and K.-H. Cho*, “Global stabilization of Boolean networks to control the heterogeneity of cellular responses”, Frontiers in Physiology, 9, 774, pp.1-17, Jul. 2018. [Abstract]
- D. Park, H.-S. Lee, J.H. Kang, S.-M. Kim, J.-R. Gong, and K.-H. Cho*, “Attractor landscape analysis of the cardiac signaling network reveals mechanism-based therapeutic strategies for heart failure”, Journal of Molecular Cell Biology, 10, 3, pp.180-194, Jun. 2018. [Abstract]
- S.-M. Choo, B. Ban, J. I. Joo, and K.-H. Cho*, “The phenotype control kernel of a biomolecular regulatory network”, BMC Systems Biology, 12, 49, pp.1-15, Apr. 2018. [Abstract]
- D. Lee and K.-H. Cho*, “Topological estimation of signal flow in complex signaling networks”, Online publication in Scientific Reports, 8, 5262, pp.1-11, Mar. 2018. [Abstract]
“This paper has been selected for Faculty 1000 Prime (https://f1000.com/prime/732915992) and evaluated by Piet van der Graaf.” - M. Choi, J. Shi, Y. Zhu, R. Yang, and K.-H. Cho*, “Network dynamics-based stratification of cancer panel for systemic prediction of anticancer drug response”, Nature Communications (IF: 12.124), 8, 1940, pp.1-12, Dec. 2017. [Abstract]
“This paper has been selected as a featured article in the interdisciplinary collection of recent research on complex systems. (www.nature.com/collections/complexitynatcomms)” - D. Shin, J. Lee, J.-R. Gong, and K.-H. Cho*, “Percolation transition of cooperative mutational effects in colorectal tumorigenesis”, Nature Communications (IF: 12.124), 8, 1270, pp.1-14, Nov. 2017. [Abstract]
“This paper has been selected as a featured article in the interdisciplinary collection of recent research on complex systems. (www.nature.com/collections/complexitynatcomms)” - J. H. Kang and K.-H. Cho*, “A novel interaction perturbation analysis reveals a comprehensive regulatory principle underlying various biochemical oscillators”, BMC Systems Biology, 11, 95, pp.1-12, Oct. 2017. [Abstract]
- J.-K. Won, S. J. Yu, C. Y. Hwang, S.-H. Cho, S.-M. Park, K. Kim, W.-M. Choi, H. Cho, E. J. Cho, J.-H. Lee, K. B. Lee, Y. J. Kim, K.-S. Suh, J.-J. Jang, C. Y. Kim, J.-H. Yoon, K.-H. Cho*, “Protein disulfide isomerase inhibition synergistically enhances the efficacy of sorafenib for hepatocellular carcinoma”, Hepatology (IF: 13.246), 66, 3, pp.855-868, Sep. 2017. [Abstract]
- J. Kim, Y. Kim, R. Nakajima, A. Shin, M. Jeong, A. H. Park, Y. Jeong, S. Jo, S. Yang, H. Park, S.-H. Cho, K.-H. Cho, I. Shim, J. H. Chung, S.-B. Paik, G. J. Augustine, and D. Kim, “Inhibitory basal ganglia inputs induce excitatory motor signals in the thalamus”, Neuron, 95, 5, pp.1181-1196, Aug. 2017. [Abstract]
- H. Chu, C. K. Chung, W. Jeong, and K.-H. Cho*, “Predicting epileptic seizures from scalp EEG based on attractor state analysis”, Computer Methods and Programs in Biomedicine, 143, pp.75-87, May. 2017. [Abstract]
“This paper has been selected for Editor’s choice in Computer Methods and Programs in Biomedicine. The articles selected by the Editor’s choice are linked on CMPB Facebook page (https://www.facebook.com/JoCMPB).” - Y. Kim, S. Choi, D. Shin, K.-H. Cho*, “Quantitative evaluation and reversion analysis of the attractor landscapes of an intracellular regulatory network for colorectal cancer”, BMC Systems Biology, 11, 45, pp.1-11, Apr. 2017. [Abstract]
- K.-H. Cho*, S. Lee, D. Kim, D. Shin, J. I. Joo, and S.-M. Park, “Cancer reversion, a renewed challenge in systems biology”, Current Opinion in Systems Biology, 2, pp.48-57, Apr. 2017. [Abstract]
- I. Yang, S. H. Jung, and K.-H. Cho*, “Self-Repairing Digital System Based on State Attractor-Convergence Inspired by the Recovery Process of a Living Cell”, IEEE Transactions on Very Large Scale Integration Systems, 25, 2, pp.648-659, Feb. 2017. [Abstract]
- J. H. Kang, H.-S. Lee, D. Park, Y.-W. Kang, S. M. Kim, J.-R. Gong, and K.-H. Cho*, “Context-independent essential regulatory interactions for apoptosis and hypertrophy in the cardiac signaling network”, Scientific Reports, 7, 34, pp.1-14, Feb. 2017. [Abstract]
- J. Kim and K.-H. Cho*, “Robustness analysis of network modularity”, IEEE Transactions on Control of Network Systems, 3, 4, pp.348-357, Dec. 2016. [Abstract]
- B. Lee and K.-H. Cho*, “Brain-inspired speech segmentation for automatic speech recognition using the speech envelope as a temporal reference”, Scientific Reports, 6, 37647, pp.1-12, Nov. 2016. [Abstract]
- S.-H. Cho, S.-M. Park, H.-S. Lee, H.-Y. Lee and K.-H. Cho*, “Attractor landscape analysis of colorectal tumorigenesis and its reversion”, BMC Systems Biology, 10, 96, pp.1-13, Oct. 2016. [Abstract]
- K.-H. Cho*, J. I. Joo, D. Shin, D. Kim, and S.-M. Park, “The reverse control of irreversible biological processes”, WIREs Systems Biology and Medicine, 8, 5, pp.366-377, Sep/Oct. 2016.

[Abstract]
“Cover Article (http://onlinelibrary.wiley.com/doi/10.1002/wsbm.1357/full) – Most biological processes have been considered to be irreversible for a long time, but some recent studies have shown the possibility of their reversion at a cellular level. How can we then understand the reversion of such biological processes? We introduce a unified conceptual framework based on the attractor landscape, a molecular phase portrait describing the dynamics of a molecular regulatory network, and the phenotype landscape, a map of phenotypes determined by the steady states of particular output molecules in the attractor landscape. In this framework, irreversible processes involve reshaping of the phenotype landscape, and the landscape reshaping causes the irreversibility of processes. We suggest reverse control by network rewiring which changes network dynamics with constant perturbation, resulting in the restoration of the original phenotype landscape. The proposed framework provides a conceptual basis for the reverse control of irreversible biological processes through network rewiring.”
“Top ten most highly accessed article in 2016 from WIREs Systems Biology and Medicine(http://wires.wiley.com/WileyCDA/WiresCollection/id-24.html).“ - S.-M. Choo and K.-H. Cho*, “An efficient algorithm for identifying primary phenotype attractors of a large-scale Boolean network”, BMC Systems Biology, 10, 95, pp.1-14, Oct. 2016 (this paper was selected as the most highly accessed paper published from BMC Systems Biology in 2016). [Abstract]
- Y.-K. Kwon, J. Kim, and K.-H. Cho*, “Dynamical Robustness Against Multiple Mutations in Signaling Networks”, IEEE/ACM Transactions on Computational Biology and Bioinformatics, 13, 5, pp. 996-1002, Sep. 2016. [Abstract]
- S.-M. Park, S.-Y. Shin, and K.-H. Cho*, “A regulated double-negative feedback decodes the temporal gradient of input stimulation in a cell signaling network”, PLoS One, 11, 9, pp.1-17, Sep. 2016. [Abstract]
- J.-Y. Hong, K. Hara, J.-W. Kim, E. Sato, E. Shim, and K.-H. Cho, “Minimal systems analysis of mitochondria-dependent apoptosis induced by cisplatin”, KJPP, 20, 4, pp.367-378, Jul. 2016. [Abstract]
- J. H. Kang, H.-S. Lee, Y.-W. Kang, and K.-H. Cho*, “Systems biological approaches to the cardiac signaling network”, Briefings in Bioinformatics, Vol. 17, Issue 3, pp.419-428, May. 2016. [Abstract]
- D. Joo, J. S. W, K.-H. Cho, S. H. Han, T. S. Min, D.-C. Yang, and C.-H. Yun*, “Biphasic activation of extracellular signal-regulated kinase (ERK) 1/2 in epidermal growth factor (EGF)-stimulated SW480 colorectal cancer cells”, BMB Reports, 49, 4, pp.220-225, Apr. 2016. [Abstract]
- S.-J. Park and K.-H. Cho*, “Power-based supervisory control theory of hybrid systems and its application to the analysis of financial crisis”, IET Control Theory & Applications, Vol. 10, Issue 7, pp.780-788, Apr. 2016. [Abstract]
- C. Hong, J. Hwang, K.-H. Cho, I. Shin*, “An efficient steady-state analysis method for large Boolean networks with high maximum node connectivity”, PLoS One, Vol. 10, Issue 12, pp.1-19, Dec. 2015. [Abstract]
- H. Chu, D. Lee, and K.-H. Cho*, “Precritical state transition dynamics in the attractor landscape of a molecular interaction network underlying colorectal tumorigenesis”, PLoS ONE, Vol. 10, Issue 10, pp.1-12, Oct. 2015. [Abstract]
- S. H. Jung, G.-Y. Kim, and K.-H. Cho*, “Evolutionary Design of Complex Digital Circuits Based on Hierarchical Module Composition and Predominant Component Prevention”, Electronics Letters, Vol. 51, Issue 20, pp. 1568-1570, Oct. 2015. [Abstract]
- H. Kang, K.-H. Cho, X. D. Zhang, T. Zeng and L. Chen*, “Inferring sequential order of somatic mutations during tumorigenesis based on Markov chain model”, IEEE/ACM Transactions on Computational Biology and Bioinformatics, Vol. 12, Issue 5, pp.1094-1103, Sep. 2015. [Abstract]
- J.-R. Kim, S.-M. Choo, H.-S. Choi, and K.-H. Cho*, “Identification of gene networks with time delayed regulation based on temporal expression profiles”, IEEE/ACM Transactions on Computational Biology and Bioinformatics, Vol. 12, Issue 5, pp.1161-1168, Sep. 2015. [Abstract]
- T.-H. Kim, N. Monsefi, J.-H. Song, A. von Kriegsheim, D. Vandamme, O. Pertz, B. N. Kholodenko, W. Kolch*, and K.-H. Cho*, “Network based identification of feedback modules that control RhoA activity and cell migration”, Journal of Molecular Cell Biology, Vol. 7, Issue 3, pp. 242-252, Jun. 2015. [Abstract]
- H.-S. Lee, M.-J. Goh, J. Kim, T. Choi, H. K. Lee, Y. J. Na, and K.-H. Cho*, “A systems-biological study on the identification of safe and effective molecular targets for the reduction of ultraviolet B-induced skin pigmentation”, Scientific Reports (Nature Publishing Group), Vol. 5: 10305, pp. 1-11, May 2015. [Abstract]
- S.-J. Park and K.-H. Cho*, “State feedback control of real-time discrete event systems with infinite states”, International Journal of Control, Vol. 88, No. 5, pp. 1078-1088, May 2015. [Abstract]
- P. Bastiaens, M. Birtwisle, N. Bluthgen, F. Bruggeman, K.-H. Cho, C. Cosentino, A. Fuente, J. Hoek, A. Kiyatkin, S. Klamt, W. Kolch, S. Legewie, P. Mendes, T. Naka, T. Santra, E. Sontag, H. Westerhoff, B. N. Kholodenko*, “Silence on the relevant literature and errors in implementation”, Nature Biotechnology, Vol. 33, No. 4, pp. 336-339, Apr. 2015. [Abstract]
- B. H. Kang, H. S. Min, Y. J. Lee, B. Choi, E. J. Kim, J. Lee, J.-R. Kim, K.-H. Cho, T. J. Kim, K. C. Jung, and S. H. Park*, “Analyses of the TCR repertoire in MHC class II-restricted innate CD4+ T cells”, Experimental & Molecular Medicine, Vol. 47, e154, Mar. 2015. [Abstract]
- K.-H. Cho*, “Editorial – Special issue celebrating the 10th anniversary of IET Systems Biology”, IET Systems Biology, Vol. 8, Issue 6, p. 243, Dec. 2014. [Abstract]
- S.-Y. Shin, T. Kim, H.-S. Lee, J. H. Kang, J. Y. Lee, K.-H. Cho* and D. H. Kim, “The switching role of β-adrenergic receptor signalling in cell survival or death decision of cardiomyocytes”, Nature Communications, Vol. 5:5777, pp. 1-13, Dec. 2014. [Abstract] [Reprint]
- T. Kaname, C.-S. Ki, N. Niikawa, G. S. Baillie, J. P. Day, K. Yamamura, T. Ohta, G. Nishimura, N. Mastuura, O.-H. Kim, Y. B. Sohn, H. W. Kim, S. Y. Cho, A.-R. Ko, J. Y. Lee, H. W. Kim, S. H. Ryu, H. Rhee, K.-S. Yang, K. Joo, J. Lee, C. H. Kim, K.-H. Cho, D. Kim, K. Yanagi, K. Naritomi, K. Yoshiura, T. Kondoh, E. Nii, H. Tonoki, M. D. Houslay, and D.-K. Jin*, “Heterozygous mutations in cyclic AMP phosphodiesterase-4D (PDE4D) and protein kinase A (PKA) provide new insights into the molecular pathology of acrodysostosis”, Cellular Signalling, Vol. 26, Issue 11, pp. 2446-2459, Nov. 2014. [Abstract]
- S. J. Yu, J.-K. Won, H. S. Ryu, W.-M. Choi, H. Cho, E.-J. Cho, J.-H. Lee, Y. J. Kim, K.-S. Suh, J.-J. Jang, C. Y. Kim, H.-S. Lee, J.-H. Yoon, and K.-H. Cho, “A novel prognostic factor for hepatocellular carcinoma: protein disulfide isomerase”, The Korean Journal of Internal Medicine, Vol. 29, Issue 5, pp. 580-587, Sep. 2014. [Abstract]
- D. Shin, I. S. Kim, J. M. Lee, S.-Y. Shin, J.-H. Lee, S. H. Baek*, and K.-H. Cho*, “The hidden switches underlying RORα-mediated circuits that critically regulate uncontrolled cell proliferation”, Journal of Molecular Cell Biology, Vol. 6, Issue 4, pp. 338-348, Aug. 2014. [Abstract]
- J. Kim D. Vandamme, J.-R. Kim, A. G. Munoz, W. Kolch, and K.-H. Cho*, “Robustness and evolvability of the human signaling network”, PLoS Computational Biology, Vol. 10, Issue 7, e1003763, Jul. 2014. [Abstract] [Reprint]
- D. Lee, M. Kim, and K.-H. Cho*, “A design principle underlying the paradoxical roles of E3 ubiquitin ligases”, Scientific Reports (Nature Publishing Group), Vol. 4: 5573, pp. 1-13, Jul. 2014. [Abstract]
- H.-S. Lee, C. Y. Hwang, S.-Y. Shin, K.-S. Kwon, and K.-H. Cho*, “MLK3 is part of a feedback mechanism that regulates different cellular responses to reactive oxygen species”, Science Signaling, Vol. 7, Issue 328, ra52, Jun. 2014. [Abstract] [Reprint] [Full text] [Summary]
“Editor’s Summary (http://stke.sciencemag.org/cgi/content/summary/sigtrans;7/328/ra52) : Balancing Responses to ROS – Low concentrations of reactive oxygen species (ROS) produced by mitochondria during oxidative metabolism symbolize fitness and signal cells to proliferate. In contrast, cell stress induces the production of high concentrations of ROS, which damage cellular proteins, lipids, and DNA and lead to cell death. Lee et al. found that two families of related kinases responded to low or high concentrations of ROS: low concentrations of ROS activated the extracellular signal–regulated kinase (ERK) family, and high concentrations of ROS activated the c-Jun N-terminal kinase (JNK) family. Mathematical modeling of the protein interaction network surrounding these two related kinase families revealed a system of feedback loops, including one involving the kinase MLK3, that balanced the relative activation of ERKs and JNKs by ROS. In cancer cells, inhibition or loss of MLK3 increased the concentration of ROS at which cells switched from activation of ERK to activation of JNK and cell death. Thus, MLK3-mediated feedback may enable cells to execute a different cellular response (proliferation or death) to different concentrations of the same stimulus (ROS). Understanding this crosstalk between kinase cascades could inform therapies that target these pathways in cancer and other diseases.” - J.-H. Song, D. J. Huels, R. A. Ridgway, O. J. Sansom, B. N. Kholodenko, W. Kolch, and K.-H. Cho*, “The APC network regulates the removal of mutated cells from colonic crypts”, Cell Reports (a new Cell press journal), Vol. 7, Issue 1, pp. 94-103, Apr. 2014. [Abstract] [Reprint]
- D. Shin and K.-H. Cho*, “Recurrent connections form a phase-locking neuronal tuner for frequency-dependent selective communication”, Scientific Reports (Nature Publishing Group), Vol. 3: 2519, pp. 1-8, Aug. 2013. [Abstract] [Reprint]
- J. Kim, S.-M. Park, and K.-H. Cho*, “Discovery of a kernel for controlling biomolecular regulatory networks”, Scientific Reports (Nature Publishing Group), Vol. 3: 2223, pp. 1-9, Jul. 2013. [Abstract] [Reprint]
- I. Yang, S. H. Jung, and K.-H. Cho*, “Self-Repairing Digital System with Unified Recovery Process Inspired by Endocrine Cellular Communication”, IEEE Transactions on Very Large Scale Integration Systems, Vol. 21, Issue 6, pp. 1027-1040, Jun. 2013. [Abstract]
- K. Noh, K. S. Shin, D. Shin, J. Y. Hwang, J. S. Kim, J. H. Jang, C. K. Chung, J. S. Kwon, and K.-H. Cho*, “Impaired coupling of local and global functional feedbacks underlies abnormal synchronization and negative symptoms of Schizophrenia”, BMC Systems Biology, Vol. 7, Issue 30, Apr. 2013. [Abstract] [Reprint]
- K.-H. Cho*, “Signaling networks, network pathology and computational chemotherapy”, Oncotarget, Vol. 4, No. 2, pp. 178-179, Feb. 2013. [Abstract]
- S. U. Chaudhary, S.-Y. Shin, D. Lee, J.-H. Song and K.-H. Cho*, “ELECANS – An integrated model development environment for multiscale cancer systems biology”, Bioinformatics, Vol. 1, No. 7, pp. 957-959, Feb. 2013. [Abstract]
“This paper has been selected to be featured in World Biomedical Frontiers (http://biomedfrontiers.org/cancer-2013-may-2-5/) because of its innovation and potential for significant impact. World Biomedical Frontiers [ISSN: 2328-0166] focuses on cutting-edge biomedical research from around the globe.” - K. Jhung, S.-H. Cho, J.-H. Jang, J. Y. Park, D. Shin, K. R. Kim, E. Lee, K.-H. Cho*, S. K. An*, “Small-world networks in individuals at ultra-high risk for psychosis and first-episode schizophrenia during a working memory task”, Neuroscience Letter, Vol. 535, pp.35-39, Feb. 2013. [Abstract]
- S.-Y. Hwang, K.-Y. Hur, J.-R. Kim, K.-H. Cho, S.-H. Kim, J.-Y. Yoo, “Biphasic RLR-IFNβ response controls the balance between antiviral immunity and cell damage”, The Journal of Immunology, Vol. 190, No. 3, pp. 1192-1200, Feb. 2013. [Abstract]
- H.-S. Choi, Y. Kim, K.-H. Cho, T. Park, “Integrative analysis of time course microarray data and DNA sequence data via log-linear models for identifying dynamic transcriptional regulatory networks”, Int. J. Data Mining and Bioinformatics., Vol. 7, No. 1, pp. 38-57, Jan. 2013. [Abstract]
- S. Kim, H. Chu, I. Yang, S. Hong, S. H. Jung, and K.-H. Cho*, “A Hierarchical Self-Repairing Architecture for Fast Fault Recovery of Digital Systems Inspired From Paralogous Gene Regulatory Circuits”, IEEE Transactions on Very Large Scale Integration Systems, Vol. 20, Issue 12, pp. 2315-2328, Dec. 2012. [Abstract]
- M. Choi, J. Shi, S. H. Jung, X. Chen, and K.-H. Cho*, “Attractor Landscape Analysis Reveals Feedback Loops in the p53 Network That Control the Cellular Response to DNA Damage”, Science Signaling, Vol. 5, Issue 251, ra83, Nov. 2012.

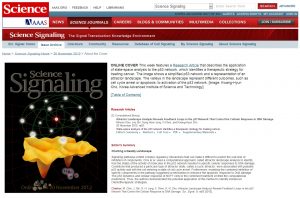

. [Abstract] [Reprint] [Full text] [Summary]
“Featured Cover Article (http://stke.sciencemag.org/content/vol5/issue251/cover.dtl) – the cover caption : This week features a Research Article that describes the application of state-space analysis to the p53 network, which identifies a therapeutic strategy for treating cancer. The image shows a simplified p53 network and a representation of an attractor landscape. The valleys in the landscape represent different outcomes, such as cell cycle arrest or apoptosis, to activation of the p53 network. [Image: Kwang-Hyun Cho, Korea Advanced Institute of Science and Technology]”
“Editor’s Summary (http://stke.sciencemag.org/cgi/content/summary/sigtrans;5/251/ra83) : Charting a Deadly Landscape – Signaling pathways exhibit complex regulatory interactions that can make it difficult to predict the outcome of inhibition of components. Choi et al. used a computational approach called attractor landscape analysis to identify how the states of the activity of molecules in the p53 network resulted in specific cellular responses to DNA damage. Conditions that produced a particular type of attractor state, called a cyclic attractor, were associated with pulsatile p53 activity and with the cell entering a state of cell cycle arrest. Furthermore, modeling the combined inhibition of specific components in the pathway suggested a mechanism to enhance the apoptotic response to DNA damage. The p53 dynamics and cellular response of MCF7 cells to the combined treatment verified the computational predictions. Thus, the authors demonstrated the potential application of this method to identify enhanced chemotherapeutic strategies.”
“Special Highlight News in Editors’ Choice of ‘Science’, 23 November 2012
(http://www.sciencemag.org/content/338/6110/twil.full) : Viewing the Cancer Landscape – The cellular signaling pathways that control cell physiology are key targets of therapeutic strategies, like those aimed at killing cancer cells. But agents that cause DNA damage in cancer cells lead to cell death through a complicated network of pathways, including positive and negative feedback loops, that control the activity of the tumor suppressor p53. To help clarify more effective strategies to promote cell death in cancer cells, Choi et al. used a computational approach in which the potential energy landscape is mapped as a three-dimensional surface in which depressions represent stable states or “attractors.” These attractors represent alternative cell fates. With a simplified network analyzed with a Boolean network model, the authors identified key network components that together could force cells to undergo apoptosis rather than proliferation or cell cycle arrest. Experiments verified that modulation of these two components effectively promoted cell death. The approach may provide a way to address complex system-level data and models to more accurately predict ways to steer cells to adopt a desired physiological state.“ - J. Kim, M. Choi, J.-R. Kim, H. Jin, V. N. Kim, and K.-H. Cho*, “The Co-regulation Mechanism of Transcription Factors in the Human Gene Regulatory Network”, Nucleic Acids Research (IF: 8.026), Vol. 40, Issue 18, pp. 8849-8861, Oct. 2012. [Abstract] [Reprint]
- D. Kim, M.-S. Kim, and K.-H. Cho*, “The core regulation module of stress-responsive regulatory networks in yeast”, Nucleic Acids Research (IF: 8.026), Vol. 40, Issue 18, pp. 8793-8802, Oct. 2012. [Abstract] [Reprint]
- C. Hong, M. Lee, D. Kim, D. Kim, K.-H. Cho and I. Shin, “A checkpoints capturing timing-robust Boolean model of the budding yeast cell cycle regulatory network”, BMC Systems Biology, Vol. 6, Issue 129, Sept. 2012. [Abstract]
- J.-Y. Hong, S.-S. Kwon, G.-H. Kim, J.-W. Kim, E. Sato, K.-H. Cho, E.-B. Shim, “Computational modeling of apoptotic signaling pathways induced by cisplatin”, BMC Systems Biology, Vol. 6, No. 122, Sept. 2012. [Abstract] [Reprint]
- J.-R. Kim and K.-H. Cho*, “The Regulatory Circuits for Hysteretic Switching in Cellular Signal Transduction Pathways”, FEBS Journal, Vol. 279 Issue 18, pp. 3329-3337, Sept. 2012. [Abstract]
- H.-W. Yang, M.-K. Shin, S. Lee, J.-R. Kim, W.-S. Park, K.-H. Cho, T. Meyer*, and W.-D. Heo*, “Cooperative activation of PI3K by Ras and Rho family small GTPases”, Molecular Cell (IF: 14.194), Vol. 47, Issue 2, pp. 281-290, Jul. 2012. [Abstract]
- J.-K. Won, H. W. Yang, S.-Y. Shin, J. H. Lee, W. D. Heo*, and K.-H. Cho*, “The Cross Regulation Between ERK and PI3K Signaling Pathways Determines the Tumoricidal Efficacy of MEK Inhibitor”, Journal of Molecular Cell Biology (IF: 13.4), Vol. 4, Issue 3, pp. 153-163, Jun. 2012.

[Abstract]
“Featured Cover Article (http://jmcb.oxfordjournals.org/content/4/3.cover-expansion) – the cover caption : A systems biological approach based on mathematical modeling and biochemical experimentation revealed that MEK inhibitor disrupts the negative feedback loops from ERK to SOS and GAB1 while activates the positive feedback loop composed of GAB1, Ras, and PI3K, which induces the bypass of ERK signal to PI3K signal and ultimately leads to the emergence of resistance to MEK inhibitor. See pages 153–163 by Won et al. for details.” - C.-Y. Dong, D. Shin, S. Joo, Y. Nam, K.-H. Cho*, “Identification of feedback loops in neural networks based on multi-step Granger causality”, Bioinformatics, Vol. 28 No. 16 pp. 2146-2153, Jun. 2012. [Abstract]
- M.-S. Kim, J.-R. Kim, D. Kim, A. D. Lander and K.-H. Cho*, “Spatiotemporal network motif reveals the biological traits of developmental gene regulatory networks in Drosophila melanogaster”, BMC Systems Biology, Vol. 6, No. 31, pp. 1-10, May 2012. [Abstract]
- C.-Y. Dong and K.-H. Cho*, “An optimally evolved connective ratio of neural networks that maximizes the occurrence of synchronized bursting behavior”, BMC Systems Biology, Vol. 6, No. 23, pp. 1-11, Mar. 2012. [Abstract]
- S.-J. Park and K.-H. Cho, “Modular nonblocking state feedback control of discrete event systems and its application to dynamic oligopolistic markets”, International Journal of Control, Vol. 84, No. 12, pp. 2046–2057, Nov. 2011. [Abstract]
- S. U. Chaudhary, S.-Y. Shin, J.-K. Won, and K.-H. Cho*, “Multi-Scale Modeling of Tumorigenesis Induced by Mitochondrial Incapacitation in Cell Death”, IEEE Transactions on Biomedical Engineering, Special Issue on “Multi-Scale Modeling and Analysis in Computational Biology and Medicine, Vol. 58, Issue 10, pp. 3028-3032, Oct. 2011. [Abstract]
- J.-R. Kim, J. Kim, Y.-K. Kwon, H.-Y. Lee, P. Heslop-Harrison, and K.-H. Cho*, “Reduction of Complex Signaling Networks to a Representative Kernel”, Science Signaling, Vol. 4, Issue 175, ra35, May 2011.
. [Abstract] [Reprint] [Full text] [Summary]
“Featured Cover Article (http://stke.sciencemag.org/content/vol4/issue175/cover.dtl) – the cover caption : This week features a Research Article that describes an algorithmic approach to simplifying complex signaling networks and then examines the properties of the nodes in those simplified networks, which may have implications for drug targeting. The image shows an artist’s rendition of a complex network overlaid with the simplified one. [Image: Chris Bickel, AAAS]”
“Editor’s Summary (http://stke.sciencemag.org/cgi/content/summary/sigtrans;4/175/ra35?ijkey=fjBWjVIPTONv6&keytype=ref&siteid=sigtrans) : Reducing Complexity – The large and complex nature of the biochemical regulatory networks that govern cell behavior provides a major challenge to the systematic analysis of cell signaling. However, most processes that reduce network complexity fail to reproduce the dynamic properties of the original network. Kim et al. describe an algorithmic approach to network reduction and simplification that preserves the dynamics of the network. They applied their approach to several networks in species from bacteria to humans, producing simplified networks called kernels. Examination of the genes represented by the kernel nodes provided insight into the evolution of these core network genes. Furthermore, the genes represented by the kernel nodes were enriched in disease-associated genes and drug targets, suggesting that this type of analysis may be therapeutically beneficial.”
“News Highlight (http://www.natureasia.com/A-IMBN/cms/preview.php?article_id=530): Stripping down biomolecular networks – Development of a technique to mathematically reduce complex biomolecular networks to their bare essentials will aid drug discovery : To facilitate the analysis of complex biomolecular networks, a team led by Kwang-Hyun Cho of the Korea Advanced Institute of Science and Technology has developed a computational algorithm that reduces them to simplified core structures, or ‘kernels’ (Fig. 1), while preserving essential dynamics and outputs1. The team’s approach should help identify new drug targets for treating human diseases. Living cells contain complex networks of interacting molecules organized into signaling pathways that regulate gene expression. However, crosstalk between signaling pathways and the existence of feedback loops complicate the modeling of network dynamics. Crosstalk can, for example, make it difficult to predict how cells will respond to environmental stimuli. Following their hunch that network kernels would be simpler to analyze without losing information, the researchers developed a kernel identification algorithm that recursively replaces local sub-networks with smaller ones that have similar dynamics. This produces a much reduced network that behaves in a similar way to the original network but whose dynamics are easier to model. The researchers have applied their algorithm to various bacterial, yeast and human signaling networks. They found that the kernel nodes identified for human signaling networks often correspond to essential genes that cause disease when mutated, implying that their approach will be useful in identifying new drug targets.”
“Focus Issue (http://stke.sciencemag.org/cgi/content/full/sigtrans;4/189/eg8): Series on Computational and Systems Biology, Sept. 2011.” - Y. Koh, K. S. Shin, J. S. Kim, J.-S. Choi, D.-H. Kang, J. H. Jang, K.-H. Cho, B. F. O’Donnell, C. K. Chung, J. S. Kwon, “An MEG study of alpha modulation in patients with schizophrenia and in subjects at high risk of developing psychosis”, Schizophrenia Research (IF: 4.458), Vol. 126, Issue 1-3, pp. 36-42, Mar. 2011. [Abstract]
- Y.-P. P. Chen*, K.-H. Cho, “The Ninth Asia Pacific Bioinformatics Conference (APBC2011) Introduction”, BMC Bioinformatics, Vol. 12, Supplement 1, pp. 1-3, Mar. 2011. [Abstract]
- T.-H. Kim, J. Kim, P. Heslop-Harrison, and K.-H. Cho*, “Evolutionary design principles and functional characteristics based on kingdom-specific network motifs”, Bioinformatics (IF: 5.039), Vol. 27, Issue 2, pp. 245-251, Jan. 2011. [Abstract]
- S.-Y. Shin, H. W. Yang, J.-R. Kim, W. D. Heo, and K.-H. Cho*, “A hidden incoherent switch regulates RCAN1 in the calcineurin–NFAT signaling network”, Journal of Cell Science (IF: 6.383), Vol. 124, Issue 1, pp. 82-90, Jan. 2011.

[Abstract]
“Featured Cover Article (http://jcs.biologists.org/content/vol124/issue1/cover.dtl) – the cover caption : A hidden incoherent regulation switch that coordinates the change in role of RCAN1 was elucidated through large-scale in silico simulations and single-cell live imaging. Kwang-Hyun Cho and colleagues found that this switch diverts negative regulation by RCAN1 to positive regulation, which leads to a biphasic response in a dose-dependent manner. See article by S.-Y. Shin et al. (pp. 82-90).”
“News Highlight (http://jcs.biologists.org/cgi/content/full/124/1/e106): RCAN1 role switch explained: The calcineurin-nuclear factor of activated T-cells (NFAT) signalling pathway controls many physiological processes including T-cell activation and cardiac-cell growth. Regulator of calcineurin (RCAN1) is a conserved regulator of this signalling network but its functional role is hotly debated because, although it inhibits calcineurin-NFAT signalling in some experimental systems, it facilitates signalling in others. To resolve this debate, Kwang-Hyun Cho, Won Do Heo, and colleagues (p. 82) have used a novel approach in which they combine in silico simulations and single-cell live imaging experimentation. The authors’ approach reveals a hidden incoherent regulation switch that is formed by crosstalk signals mediated through extracellular signal-regulated protein kinase and glycogen synthase kinase-3. This switch, they report, diverts negative regulation by RCAN1 to positive regulation, which leads to a biphasic response in a dose-dependent manner. Specifically, RCAN1 functions as an inhibitor when its levels are low but as a facilitator when its levels are high. The discovery of this incoherent regulation switch, which can induce apparently opposite responses depending on conditions, provides an explanation for the different roles of RCAN1 in calcineurin-NFAT signaling that have been suggested in previous studies, conclude the authors. By Dr. Jane Bradbury” - S.-Y. Shin, O. Rath, A. Zebisch, S.-M. Choo, W. Kolch, and K.-H. Cho*, “Functional roles of multiple feedback loops in ERK and Wnt signaling pathways that regulate epithelial-mesenchymal transition”, Cancer Research (IF: 7.543), Vol. 70, Issue 17, pp. 6715-6724, Sept. 2010. [Abstract]
- P. J. Murray, J.-W. Kang, G. R. Mirams, S.-Y. Shin, H. M. Byrne, P. K. Maini, and K.-H. Cho*, “Modelling spatially regulated beta-catenin dynamics and invasion in intestinal crypts”, Biophysical Journal (IF: 4.683), Vol. 99, Issue 3, pp. 716-725, Aug. 2010. [Abstract]
“This paper has been selected for Faculty of 1000 Biology as a ‘must read’ paper and evaluated by Stephen Lockett: see http://f1000biology.com/article/id/4811956 .”
“It has been also selected as a highly recommended paper in BRIC Hanbitsa: see http://bric.postech.ac.kr/myboard/read.php?Board=hbs_treatise&id=19504&ttype=2&idauthorid=11805 .” - J. Gim, H.-S. Kim, J. Kim, M. Choi, J.-R. Kim, Y. J. Chung, and K.-H. Cho*, “A system-level investigation into the cellular toxic response mechanism mediated by AhR signal transduction pathway”, Bioinformatics (IF: 5.039), Vol. 26, No. 17, pp. 2169-2175, Jul. 2010. [Abstract]
- M. Eshaghi, J. H. Lee, L. Zhu, S. Y. Poon, J. Li, K.-H. Cho, Z. Chu, R. K. M. Karuturi, and J. Liu, “Genomic binding profiling of the fission yeast stress-activated MAPK Sty1 and the bZIP transcriptional activator Atf1 in response to H2O2”, PLoS ONE (IF: 4.351), Vol. 5, Issue 7, e11620(1-16), Jul. 2010. [Abstract]
- M.-S. Kim, J.-R. Kim, and K.-H. Cho*, “Dynamic network rewiring determines temporal regulatory functions in the Drosophila melanogaster development processes”, BioEssays (IF: 5.402), Vol. 32, Issue 6, pp. 505-513, Jun. 2010.

[Abstract]
“Cover Photograph (http://www3.interscience.wiley.com/journal/123443528/abstract): Resolving developmental genetics in the fourth dimension: an illustration of the principle of dynamic network motifs in Drosophila development. Hitherto largely considered in terms of time-invariant networks, drosophila development is viewed in the article by Man-Sun Kim, Jeong-Rae Kim, and Kwang-Hyun Cho as the result of networks of gene interactions that change during the course of development. Using this paradigm, pivotal developmental events can be correlated with particular changes from one constellation of gene interactions to a different one, and the method allows the identification of certain network motifs that are not identifiable using static approaches. Features article: Dynamic network rewiring determines temporal regulatory functions in the Drosophila melanogaster development processes, see pages 505 – 513.” - J.-S. Kim, N.V. Valeyev, I. Postlethwaite, P. Heslop-Harrison, K.-H. Cho*, and D.G. Bates, “Analysis and Extension of a Biochemical Network Model using Robust Control Theory”, International Journal of Robust and Nonlinear Control, Vol. 20, Issue 9, pp. 1017-1026, Jun. 2010. [Abstract]
- J.-R. Kim, D. Shin, S. H. Jung, P. Heslop-Harrison, and K.-H. Cho*, “A design principle underlying the synchronization of oscillations in cellular systems”, Journal of Cell Science (IF: 6.383), Vol. 123, Issue 4, pp. 537-543, Feb. 2010. [Abstract]
- C.-Y. Dong, T.-W. Yoon, D. G. Bates, and K.-H. Cho*, “Identification of feedback loops embedded in cellular circuits by investigating non-causal impulse response components”, Journal of Mathematical Biology, Vol. 60, No. 2, pp. 285-312, Feb. 2010. [Abstract]
- D. Kim, W. Kolch and K.-H. Cho*, “Multiple roles of the NF-kappaB signaling pathway regulated by coupled negative feedback circuits”, FASEB Journal (IF: 7.049), Vol. 23, Issue 9, pp. 2796-2802, Sept. 2009. [Abstract]
- C. H. Seo, J.-R. Kim, M.-S. Kim, and K.-H. Cho*, “Hub genes with positive feedbacks function as master switches in developmental gene regulatory networks”, Bioinformatics (IF: 5.039), Vol. 25, No. 15, pp. 1898-1904, Aug. 2009. [Abstract]
- C.-Y. Dong, J. Lim,Y. Nam, and K.-H. Cho*, “Systematic analysis of synchronized oscillatory neuronal networks reveals the enrichment of coupled direct and indirect feedback motifs”, Bioinformatics (IF: 5.039), Vol. 25, No. 13, pp. 1680-1685, Jul. 2009. [Abstract]
- S.-J. Park and K.-H. Cho*, “Delay-coobservability and its algebraic properties for decentralized supervisory control of discrete event systems with communication delays”, Automatica, Vol. 45, No. 5, pp. 1252-1259, May. 2009. [Abstract]
- S.-Y. Shin, O. Rath, S.-M. Choo, F. Fee, B. McFerran, W. Kolch, and K.-H. Cho*, “Positive- and negative-feedback regulations coordinate the dynamic behavior of the Ras-Raf-MEK-ERK signal transduction pathway”, Journal of Cell Science (IF: 6.383), Vol. 122, Issue 3, pp. 425-435, Feb. 2009.

[Abstract]
“Cover Paper (http://jcs.biologists.org/content/vol122/issue3/cover.shtml): The hidden dynamics of the positive- and negative-feedback mechanisms and the functional role of RKIP in the ERK pathway are elucidated through a combined study of biochemical in vitro experiments and in silico simulations.” - S. Kim, M. Choi, and K.-H. Cho*, “Identifying the target mRNAs of microRNAs in colorectal cancer”, Computational Biology and Chemistry, Vol. 33, Issue 1, pp. 94-99, Feb. 2009. [Abstract]
- S.-J. Park and K.-H. Cho*, “Supervisory control for fault-tolerant scheduling of real-time multiprocessor systems with aperiodic tasks”, International Journal of Control, Vol. 82, No. 2, pp. 217-227, Feb. 2009. [Abstract]
- D. Kim, Y.-K. Kwon, and K.-H. Cho*, “The biphasic behavior of incoherent feed-forward loops in biomolecular regulatory networks”, BioEssays (IF: 5.402), Vol. 30, Issue 11-12, pp. 1204-1211, Nov. 2008. [Abstract]
- Y.-K. Kwon and K.-H. Cho*, “Coherent coupling of feedback loops: A design principle of cell signaling networks”, Bioinformatics (IF: 5.039), Vol. 24, No. 17, pp. 1926-1932, Sept. 2008. [Abstract]
“This paper has been selected for Faculty of 1000 Biology (http://www.f1000biology.com) and evaluated by Andreas Wagner: see http://www.f1000biology.com/article/id/1116696.“ - S.-J. Park and K.-H. Cho*, “Real-Time Preemptive Scheduling of Sporadic Tasks Based on Supervisory Control of Discrete Event Systems”, Information Sciences (IF: 3.291), Vol. 178, Issue 17, pp. 3393-3401, Sept. 2008. [Abstract]
- J. Kim, T.-G. Kim, S. H. Jung, J.-R. Kim, T. Park, P. Heslop-Harrison, and K.-H. Cho*, “Evolutionary design principles of modules that control cellular differentiation: Consequences for hysteresis and multistationarity”, Bioinformatics (IF: 5.039), Vol. 24, No. 13, pp. 1516-1522, Jul. 2008. [Abstract]
- T. Millat*, S. N. Sreenath, R. P. Soebiyanto, J. Avva, K.-H. Cho, and O. Wolkenhauer, “The role of dynamic stimulation pattern in the analysis of bistable intracellular networks”, BioSystems, Vol. 92, No. 3, pp. 270-281, Jun. 2008. [Abstract]
- H.-S. Choi, J.-R. Kim, S.-W. Lee, and K.-H. Cho*, “Why have serine/threonine/tyrosine kinases been evolutionarily selected in eukaryotic signaling cascades?”, Computational Biology and Chemistry, Vol. 32, Issue 3, pp. 218-221, Jun. 2008. [Abstract]
- S.-Y. Shin, J. M. Yang, S.-M. Choo, K.-S. Kwon, and K.-H. Cho*, “System-Level Investigation into the Regulatory Mechanism of the Calcineurin/NFAT Signaling Pathway”, Cellular Signalling (IF: 4.305), Vol. 20, Issue 6, pp. 1117-1124, Jun. 2008. [Abstract]
- K.-I. Goh, B. Kahng, and K.-H. Cho*, “Sustained oscillations in extended genetic oscillatory systems”, Biophysical Journal (IF: 4.683), Vol. 94, Issue 11, pp. 4270-4276, Jun. 2008. [Abstract]
- J. Kim, D. G. Bates, I. Postlethwaite, P. Heslop-Harrison, and K.-H. Cho*, “Linear time-varying models can reveal nonlinear interactions of biomolecular regulatory networks using multiple time-series data”, Bioinformatics (IF: 5.039), Vol. 24, No. 10, pp. 1286-1292, May 2008. [Abstract]
- T.-H. Kim, S.-Y. Shin, S.-M. Choo, and K.-H. Cho*, “Dynamical analysis of the calcium signaling pathway in cardiac myocytes based on logarithmic sensitivity analysis”, Biotechnology Journal, Vol. 3, No. 5, pp. 639-647, May 2008. [Abstract]
- Y.-K. Kwon and K.-H. Cho*, “Quantitative analysis of robustness and fragility in biological networks based on feedback dynamics”, Bioinformatics (IF: 5.039), Vol. 24, No. 7, pp. 987-994, Apr. 2008. [Abstract]
- S.-J. Park and K.-H. Cho*, “Nonblocking supervisory control of timed discrete event systems under communication delays: The existence conditions”, Automatica, Vol. 44, Issue 4, pp. 1011-1019, Apr. 2008. [Abstract]
- S.-J. Park and K.-H. Cho*, “Robust supervisory control of timed discrete event systems under partial observation based on eligible time bounds: The existence conditions”, Automatica, Vol. 44, Issue 3, pp. 875-881, Mar. 2008. [Abstract]
- J.-R. Kim, Y. Yoon, and K.-H. Cho*, “Coupled feedback loops form dynamic motifs of cellular networks”, Biophysical Journal (IF: 4.683), Vol. 94, Issue 2, pp. 359-365, Jan. 2008. [Abstract]
- T.-H. Kim, S. H. Jung, and K.-H. Cho*, “Investigations into the design principles in the chemotactic behavior of Escherichia coli”, BioSystems, Vol. 91, Issue 1, pp. 171-182, Jan. 2008. [Abstract]
- Y.-K. Kwon and K.-H. Cho*, “Analysis of Feedback Loops and Robustness in Network Evolution Based on Boolean Models”, BMC Bioinformatics (IF: 3.493), Vol. 8, No. 430, pp. 1-9, Nov. 2007. [Abstract]
- Y.-K. Kwon, S. S. Choi, and K.-H. Cho*, “Investigations into the relationship between feedback loops and functional importance of a signal transduction network based on Boolean network modeling”, BMC Bioinformatics (IF: 3.493), Vol. 8, No. 384, pp. 1-9, Oct. 2007. [Abstract]
- T.-H. Kim, S. H. Jung, and K.-H. Cho*, “Interlinked mutual inhibitory positive feedbacks induce robust cellular memory effects”, FEBS Letters (IF: 3.541), Vol. 581, Issue 25, pp. 4899-4904, Oct. 2007. [Abstract]
- M. Kim, S. Baek, S. H. Jung, and K.-H. Cho*, “Dynamical characteristics of bacteria clustering by self-generated attractants”, Computational Biology and Chemistry, Vol. 31, Issue 5-6, pp. 328-334, Oct. 2007. [Abstract]
- S. Han, Y. Yoon, and K.-H. Cho*, “Inferring biomolecular interaction networks based on convex optimization”, Computational Biology and Chemistry, Vol. 31, Issue 5-6, pp. 347-354, Oct. 2007. [Abstract]
- S. Kim, J. Kim, and K.-H. Cho*, “Inferring gene regulatory networks from temporal expression profiles under time-delay and noise”, Computational Biology and Chemistry, Vol. 31, Issue 4, pp. 239-245, Aug. 2007. [Abstract]
- D. Kim, O. Rath, W. Kolch, and K.-H. Cho*, “A hidden oncogenic positive feedback loop caused by crosstalk between Wnt and ERK pathways”, Oncogene (IF: 7.216), Vol. 26, Issue 31, pp. 4571-4579, Jul. 2007. [Abstract]
- J.-R. Kim, W.-S. Bae, Y. Yoon, and K.-H. Cho*, “Topological difference of core regulatory networks induces different entrainment characteristics of plant and animal circadian clocks”, Biophysical Journal (IF: 4.683), Vol. 93, Issue 1, pp. L01-L03, Jul. 2007. [Abstract]
- H.-S. Choi, S. Han, H. Yokota, and K.-H. Cho*, “Coupled positive feedbacks provoke slow induction plus fast switching in apoptosis”, FEBS Letters (IF: 3.541), Vol. 581, Issue 14, pp. 2684-2690, Jun. 2007. [Abstract]
- K.-H. Cho*, H.-S. Choi, and S.-M. Choo, “Unraveling the functional interaction structure of a biomolecular network through alternate perturbation of initial conditions”, Journal of Biochemical and Biophysical Methods, Vol. 70, Issue 4, pp. 701-707, Jun. 2007. [Abstract]
- K.-H. Cho*, S.-M. Choo, S. H. Jung, J.-R. Kim, H.-S. Choi, and J. Kim, “Reverse Engineering of Gene Regulatory Networks”, IET Systems Biology, Vol. 1, No. 3, pp. 149-163, May 2007. [Abstract]
“This paper is ranked No. 5 most downloaded IET Systems Biology papers by users of the IEEE Xplore database in 2008 from the hundreds of papers that the journal IET Systems Biology has published since its launch in 2004, receiving 149 full text downloads only in 2008.” - Y.-K. Kwon and K.-H. Cho*, “Boolean dynamics of biological networks with multiple coupled feedback loops”, Biophysical Journal (IF: 4.683), Vol. 92, Issue 8, pp. 2975-2981, Apr. 2007. [Abstract]
- S.-J. Park and K.-H. Cho*, “Decentralized supervisory control of discrete event systems with communication delays based on conjunctive and permissive decision structures”, Automatica, Vol. 43, Issue 4, pp. 738-743, Apr. 2007. [Abstract]
- S.-J. Park and K.-H. Cho*, “Supervisory control of discrete event systems with communication delays and partial observations”, Systems & Control Letters, Vol. 56, Issue 2, pp. 106-112, Feb. 2007. [Abstract]
- S.-J. Park and K.-H. Cho*, “Decentralized supervisory control of nondeterministic discrete event systems: The existence condition of a robust and nonblocking supervisor”, Automatica, Vol. 43, Issue 2, pp. 377-383, Feb. 2007. [Abstract]
- J. Kim, D. G. Bates, I. Postlethwaite, P. Heslop-Harrison, and K.-H. Cho*, “Least-squares methods for identifying biochemical regulatory networks from noisy measurements”, BMC Bioinformatics (IF: 3.493), Vol. 8, No. 8, pp. 1-15, Jan. 2007. [Abstract]
- D. Kim, Y.-K. Kwon, and K.-H. Cho*, “Coupled positive and negative feedback circuits form an essential building block of cellular signaling pathways”, BioEssays (IF: 5.402), Vol. 29, Issue 1, pp. 85-90, Jan. 2007. [Abstract]
- J.-R. Kim and K.-H. Cho*, “The multi-step phosphorelay mechanism of unorthodox two-component systems in E. coli realizes ultrasensitivity to stimuli while maintaining robustness to noises”, Computational Biology and Chemistry, Vol. 30, Issue 6, pp. 438-444, Dec. 2006. [Abstract]
- H. E. Assmus*, R. Herwig, K.-H. Cho, and O. Wolkenhauer, “Dynamics of biological systems: Role of systems biology in medical research”, Expert Review of Molecular Diagnostics (IF: 3.693), Vol. 6, No. 6, pp. 891-902, Nov. 2006. [Abstract]
- S.-Y. Shin, S.-M. Choo, D. Kim, S. J. Baek, O. Wolkenhauer, and K.-H. Cho*, “Switching feedback mechanisms realize the dual role of MCIP in the regulation of calcineurin activity”, FEBS Letters (IF: 3.541), Vol. 580, Issue 25, pp. 5965-5973, Oct. 2006. [Abstract]
- D.-Y. Cho, K.-H. Cho*,and B.-T. Zhang, “Identification of biochemical networks by S-tree based genetic programming”, Bioinformatics (IF: 5.039), Vol. 22, No. 13, pp. 1631-1640, Jul. 2006. [Abstract]
- S.-J. Park, K.-H. Cho,and J.-T. Lim*, “Nonblocking supervisory control under bounded time constraints based on nondeterministic timed transition models”, IEE Proceedings – Control Theory and Applications, Vol. 153, No. 4, pp. 419-426, Jul. 2006. [Abstract]
- K.-H. Cho*, S. Baek, and M.-H. Sung, “Wnt pathway mutations selected by optimal β-catenin signaling for tumorigenesis”, FEBS Letters (IF: 3.541), Vol. 580, Issue 15, pp. 3665-3670, Jun. 2006. [Abstract]
- K.-H. Cho*, J.-R. Kim, S. Baek, H.-S. Choi, and S.-M. Choo, “Inferring biomolecular regulatory networks from phase portraits of time-series expression profiles”, FEBS Letters (IF: 3.541), Vol. 580, Issue 14, pp. 3511-3518, Jun. 2006. [Abstract]
- S.-J. Park and K.-H. Cho*, “Delay-robust supervisory control of discrete event systems with bounded communication delays”, IEEE Transactions on Automatic Control, Vol. 51, No. 5, pp. 911-915, May 2006. [Abstract]
- S.-J. Park and K.-H. Cho*, “Supervisory control of timed discrete event systems under partial observation based on activity models and eligible time bounds”, Systems & Control Letters, Vol. 55, Issue 5, pp. 407-413, May 2006. [Abstract]
- M. Ullah, H. Schmidt, K.-H. Cho, and O. Wolkenhauer*, “Deterministic modelling and stochastic simulation of biochemical pathways using MATLAB”, IEE Proceedings – Systems Biology, Vol. 153, No. 2, pp. 53-60, Mar. 2006. [Abstract]
- S. G. Park, T. Lee, H. Y. Kang, K. Park, K.-H. Cho*, and G. Jung, “The influence of the signal dynamics of activated form of IKK on NF-κB and anti-apoptotic gene expressions: A systems biology approach”, FEBS Letters (IF: 3.541), Vol. 580, Issue 3, pp. 822-830, Feb. 2006. [Abstract]
- S.-J. Park, M.-S. Lee, S.-Y. Shin, K.-H. Cho*, J.-T. Lim, B.-S. Cho, Y.-H. Jei, M.-K. Kim, and C.-H. Park, “Run-to-run overlay control of steppers in semiconductor manufacturing systems based on history data analysis and neural network modeling”, IEEE Transactions on Semiconductor Manufacturing, Vol. 18, No. 4, pp. 605-613, Nov. 2005. [Abstract]
- K.-H. Cho*, S.-J. Park, E.-H. Jung, S.-W. Shin, and H.-H. Lee, “End-to-end rate-based congestion control using EWMA for multicast services in IP based networks”, IEE Proceedings – Communications, Vol. 152, No. 5, pp. 668-672, Oct. 2005. [Abstract]
- K.-H. Cho*, S.-M. Choo, P. Wellstead, and O. Wolkenhauer, “A Unified framework for unraveling the functional interaction structure of a biomolecular network based on stimulus-response experimental data”, FEBS Letters (IF: 3.541), Vol. 579, Issue 20, pp. 4520-4528, Aug. 2005. [Abstract]
- K.-H. Cho*, S.-Y. Shin, and S.-M. Choo, “Unravelling the functional interaction structure of a cellular network from temporal slope information of experimental data”, FEBS Journal (IF: 3.396), Vol. 272, Issue 15, pp. 3950-3959, Aug. 2005. [Abstract]
- J.-M. Lim and K.-H. Cho*, “Prediction of gene expression levels and the role of cis-acting elements in age-related cataract by applying promoter-based modeling approach”, Biotechnology Progress, Vol. 21, No. 4, pp. 1032-1037, Jul. 2005. [Abstract]
- O. Wolkenhauer*, S. N. Sreenath, P. Wellstead, M. Ullah, and K.-H. Cho, “A systems- and signal-oriented approach to intracellular dynamics”, Biochemical Society Transactions (IF: 3.447), Vol. 33, No. 3, pp. 507-515, Jun. 2005. [Abstract]
- K.-H. Cho*, K. H. Johansson, and O. Wolkenhauer, “A hybrid systems framework for cellular processes”, BioSystems, Vol. 80, Issue 3, pp. 273-282, Jun. 2005. [Abstract]
- H. Schmidt, K.-H. Cho*, and E. W. Jacobsen, “Identification of small scale biochemical networks based on general type system perturbations”, FEBS Journal (IF: 3.396), Vol. 272, Issue 9, pp. 2141-2151, May 2005. [Abstract]
- C. S. Moller, F. Klawonn, K.-H. Cho*, H. Yin, and O. Wolkenhauer, “Clustering of unevenly sampled gene expression time-series data”, Fuzzy Sets and Systems, Vol. 152, Issue 1, pp. 49-66, May 2005. [Abstract]
- K.-H. Cho* and O. Wolkenhauer, “Systems biology: Discovering the dynamic behavior of biochemical networks”, BioSystems Review, Vol. 1, No. 1, pp. 9-17, Mar. 2005. [Abstract]
- O. Wolkenhauer*, M. Ullah, P. Wellstead, and K.-H. Cho, “The dynamic systems approach to control and regulation of intracellular networks”, FEBS Letters (IF: 3.541), Vol. 579, Issue 8, pp. 1846-1853, Mar. 2005. [Abstract]
- Z. Zi, K.-H. Cho*, M.-H. Sung, X. Xia, J. Zheng, and Z. Sun, “In silico identification of the key components and steps in IFN-γ induced JAK-STAT signaling pathway”, FEBS Letters (IF: 3.541), Vol. 579, Issue 5, pp. 1101-1108, Feb. 2005. [Abstract]
- K.-O. Nam, H. Kang, S.-M. Shin, K.-H. Cho, B. Kwon, B. S. Kwon, S.-J. Kim, and H.-W. Lee*, “Cross-linking of 4-1BB activates TCR-signaling pathways in CD8+ T lymphocytes”, Journal of Immunology (IF: 6.068), Vol. 174, No. 4, pp. 1898-1905, Feb. 2005. [Abstract]
- O. Wolkenhauer*, M. Ullah, W. Kolch, and K.-H. Cho, “Modelling and simulation of intracellular dynamics: Choosing an appropriate framework”, IEEE Trans. on NanoBioscience, Vol. 3, No. 3, pp. 200-207, Sep. 2004. [Abstract]
- O. Wolkenhauer, B. K. Ghosh, and K.-H. Cho*, “Control and coordination in biochemical networks”, IEEE Control Systems Magazine, Vol. 24, No. 4, pp. 30-34, Aug. 2004. [Abstract]
- Z. Kutalik, K.-H. Cho, and O. Wolkenhauer*, “Optimal sampling time selection for parameter estimation in signal transduction pathway modeling”, BioSystems, Vol. 75, Issue 1-3, pp. 43-55, Jul. 2004. [Abstract]
- S.-J. Park, K.-H. Cho, and J.-T. Lim*, “Supervisory control of real-time discrete event systems under bounded time constraints”, IEE Proceedings – Control Theory and Applications, Vol. 151, No. 3, pp. 347-352, May 2004. [Abstract]
- Z. Kutalik, J. Inwald, S. V. Gordon, R. G. Hewinson, P. Butcher, J. Hinds, K.-H. Cho, and O. Wolkenhauer*, “Advanced significance analysis of microarray data based on weighted resampling: A comparative study and application to gene deletions in Mycobacterium bovis”, Bioinformatics (IF: 5.039), Vol. 20, No. 3, pp. 357-363, Feb. 2004. [Abstract]
- K.-H. Cho*, S.-Y. Shin, W. Kolch, and O. Wolkenhauer, “Experimental design in systems biology based on parameter sensitivity analysis with Monte Carlo method: A case study for the TNFα mediated NF-κB signal transduction pathway”, SIMULATION: Transactions of The Society for Modeling and Simulation International, Vol. 79, Issue 12, pp. 726-739, Dec. 2003. [Abstract]
- K.-H. Cho* and O. Wolkenhauer, “Analysis and modelling of signal transduction pathways in systems biology”, Biochemical Society Transactions (IF: 3.447), Vol. 31, No. 6, pp. 1503-1509, Dec. 2003. [Abstract]
- K.-H. Cho*, S.-Y. Shin, H.-W. Lee, and O. Wolkenhauer, “Investigations into the analysis and modeling of the TNFα mediated NF-κB signaling pathway”, Genome Research (IF: 11.342), Vol. 13, No. 11, pp. 2413-2422, Nov. 2003. [Abstract]
- J. Nunez-Garcia, Z. Kutalik, K.-H. Cho, and O. Wolkenhauer*, “Level sets and minimum volume sets of probability density functions”, International Journal of Approximate Reasoning, Vol. 34, No. 1, pp. 25-47, Sept. 2003. [Abstract]
- O. Wolkenhauer, H. Kitano, and K.-H. Cho*, “Systems biology: Looking at opportunities and challenges in applying systems theory to molecular and cell biology”, IEEE Control Systems Magazine, Vol. 23, No. 4, pp. 38-48, Aug. 2003. [Abstract]
- S.-G. Kim, K.-H. Cho, and J.-T. Lim*, “Hierarchical supervisory control of discrete event systems based on H-observability”, IEE Proceedings – Control Theory and Applications, Vol. 150, No. 2, pp. 179-182, Mar. 2003. [Abstract]
- J. Nunez-Garcia, V. Mersinias, K.-H. Cho, C. Smith, and O. Wolkenhauer*, “The statistical distribution of the intensity of pixels within spots of DNA microarrays: What is the appropriate single-valued representative?”, Applied Bioinformatics, Vol. 2, No. 4, pp. 229-239, 2003. [Abstract]
- C. S. Moller, K.-H. Cho, and O. Wolkenhauer*, “Microarray data clustering based on temporal variation: FCV with TSD preclustering”, Applied Bioinformatics, Vol. 2, No. 1, pp. 35-45, 2003. [Abstract]
- K.-H. Cho* and S.-W. Shin, “Consolidation algorithm based on adaptive dynamic threshold for point-to-multipoint connections in ATM networks”, IEICE Trans. on Communications, Vol. E85-B, No. 3, pp. 686-688, Mar. 2002. [Abstract]
- K.-H. Cho*, B.-H. Kim, and K.-S. Park, “Case study on rate-based traffic control of industrial networks employing LonWorks”, International Journal of Systems Science, Vol. 33, No. 3, pp. 161-164, Feb. 2002. [Abstract]
- K.-H. Cho* and J.-T. Lim, “Multiagent supervisory control for anti-fault-propagation in serial production systems”, IEEE Trans. on Industrial Electronics (IF: 4.678), Vol. 48, No. 2, pp. 460-466, Apr. 2001. [Abstract]
- K.-H. Cho* and J.-T. Lim, “Fault-tolerant supervisory control under C, D observability and its application”, International Journal of Systems Science, Vol. 31, No. 12, pp. 1573-1583, Dec. 2000. [Abstract]
- K.-H. Cho*, M.-S. Lee, and J.-T. Lim, “Supervisory control for optimal route guidance in IVHSs based on hybrid network models”, International Journal of Systems Science, Vol. 31, No. 9, pp. 1107-1113, Sept. 2000. [Abstract]
- K.-H. Cho* and J.-T. Lim, “Unobservable fault-propagation control of serial production systems based on discrete event models”, IEE Proceedings – Control Theory and Applications, Vol. 147, No. 4, pp. 381-386, Jul. 2000. [Abstract]
- K.-H. Cho and J.-T. Lim*, “On-line tracing supervisory control of discrete event dynamic systems based on outlooking”, Automatica, Vol. 35, No. 10, pp. 1725-1729, Oct. 1999. [Abstract]
- K.-H. Cho and J.-T. Lim*, “Layered optimal supervisory control of discrete event dynamic systems”, International Journal of Systems Science, Vol. 30, No. 4, pp. 395-405, Apr. 1999. [Abstract]
- K.-H. Cho and J.-T. Lim*, “Mixed centralized/decentralized supervisory control of discrete event dynamic systems”, Automatica, Vol. 35, No. 1, pp. 121-128, Jan. 1999. [Abstract]
- K.-H. Cho and J.-T. Lim*, “Supervisory rate-based flow control of ATM networks for ABR services”, IEICE Trans. on Communications, Vol. E81-B, No. 6, pp. 1269-1271, Jun. 1998. [Abstract]
- K.-H. Cho and J.-T. Lim*, “Synthesis of fault-tolerant supervisor for automated manufacturing systems: a case study on photolithographic process”, IEEE Trans. on Robotics and Automation, Vol. 14, No. 2, pp. 348-351, Apr. 1998. [Abstract]
- K.-H. Cho and J.-T. Lim*, “A study on stability analysis of discrete event dynamic systems”, IEICE Trans. on Information and Systems, Vol. E80-D, No. 12, pp. 1149-1154, Dec. 1997. [Abstract]
- K.-H. Cho and J.-T. Lim*, “Fault-tolerant supervisory control of discrete event dynamical systems”, International Journal of Systems Science, Vol. 28, No. 10, pp. 1001-1009, Jul. 1997. [Abstract]
- K.-H. Cho and J.-T. Lim*, “Stability and robustness of discrete event dynamic systems”, International Journal of Systems Science, Vol. 28, No. 7, pp. 691-703, Jul. 1997. [Abstract]
- K.-H. Cho and J.-T. Lim*, “Nonhomogeneous decentralized supervisory control of discrete event dynamic systems”, IEICE Trans. on Information and Systems, Vol. E80-D, No. 5, pp. 605-607, May 1997. [Abstract]
- K.-H. Cho and J.-T. Lim*, “Failure diagnosis and fault tolerant supervisory control system”, IEICE Trans. on Information and Systems, Vol. E79-D, No. 9, pp. 1223-1231, Sept. 1996. [Abstract]
- S. H. Jung*, K.-H. Cho, T. G. Kim, K. H. Park, and J.-T. Lim, “Design of flexible PID-plus bang-bang controller with neural network predictive model”, IEICE Trans. on Information and Systems, Vol. E79-D, No. 4, pp. 357-362, Apr. 1996. [Abstract]
- K.-H. Cho and J.-T. Lim*, “On Lagrangian approach to mixed H2/H∞ control problem: The state feedback case”, Journal of Electrical Engineering and Information Science, Vol. 1, No. 1, pp. 29-38, Mar. 1996. [Abstract]
- S. H. Jung*, K.-H. Cho, T. G. Kim, and K. H. Park, “Defuzzification method for multishaped output fuzzy sets”, Electronics Letters, Vol. 30, No. 9, pp. 740-742, Apr. 1994. [Abstract]